Overview
-

Merits of tree identification: When comparing a query sequence with reference sequences, phyloBARCODER can identify closer sequence even sharing the same BLAST identity (result674_Tree_vs_similarity.zip).
As input, users upload a set of sequences obtained from environmental DNA or metabarcoding samples (referred to here as anonymous sequences). Users can also utilize custom reference sequences as their own database. As an expected outcome, even when BLAST identity is the same, the tool may select a sequence that is phylogenetically closer (result674_Tree_vs_similarity.zip).
(A) Species Identification
-
Here, we use 12S rRNA eDNA sequences, as an example of anonymous sequences for uploading. Yu et al. (2022) amplified those sequences by using MiFish primers.
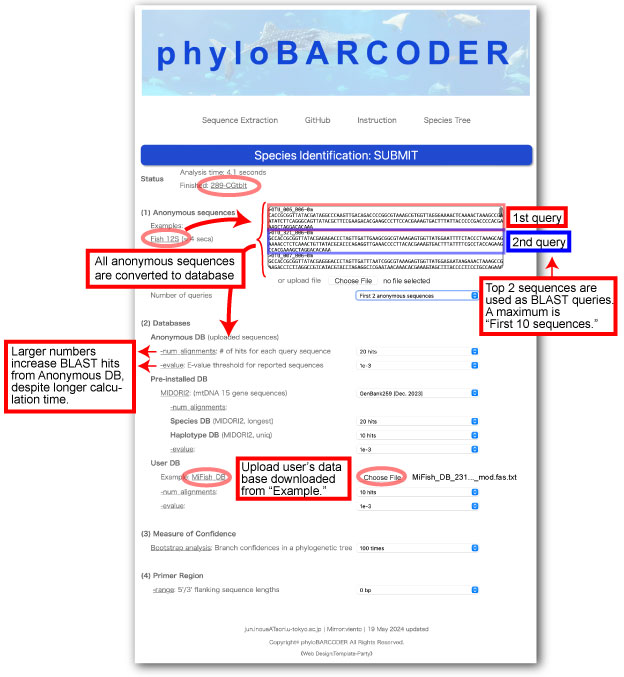
The user copy & pastes anonymous eDNA sequences obtained from the link, Fish 12S, to the text box.
To identify species names for 2 OTUs simultaneously, Number of queries is set as First 2 sequences.
- Anonymous sequences
- KS1815-B06-0m_ASV.fasta.txt
- Raw data
- KS1815-B06-0m_S50_R1_001.fastq.gz
- KS1815-B06-0m_S50_R2_001.fastq.gz
- (Yu et al. 2021) Example: Copepod analysis
- Reference sequence
- Metridia_pacifica_lucens_MZGdb_Selected.txt
- (MetaZooGene Atlas & Database, 11 Oct 2023)
Number of queries: This box has a maximum of “First 10 sequences.” When “First 2 sequences” are selected, only the first 2 are used as queries for BLAST searches. However, all anonymous sequences including 2 query sequences are converted to “Anonymous DB.” Therefore, anonymous sequences having close matches with the first 2 sequences are included in the multiple sequence alignment. The remaining anonymous sequences (not selected by BLAST) are not included in the alignment.
-num_alignments or -evalue options: For all databases, those options clarify species identification by adjusting sequence members included in resultant alignments.
- For Anonymous DB, these numbers increase or decrease the number of BLAST hits in order to clarify anonymous sequences belonging to the same groups of query sequences.
-For Species DB (Pre-installed DB), those numbers can identify appropriate root sequences for focal species/groups in resultant phylogenetic trees, and for Haplotype DB, they delineate species/group or population groups clearer with different sequences of the same reference species.
- For User DB, those numbers increase or decrease related sequences of focal species and relatives.
- If “- num_alignments” is set as “0 (not used)”, the database is not used.
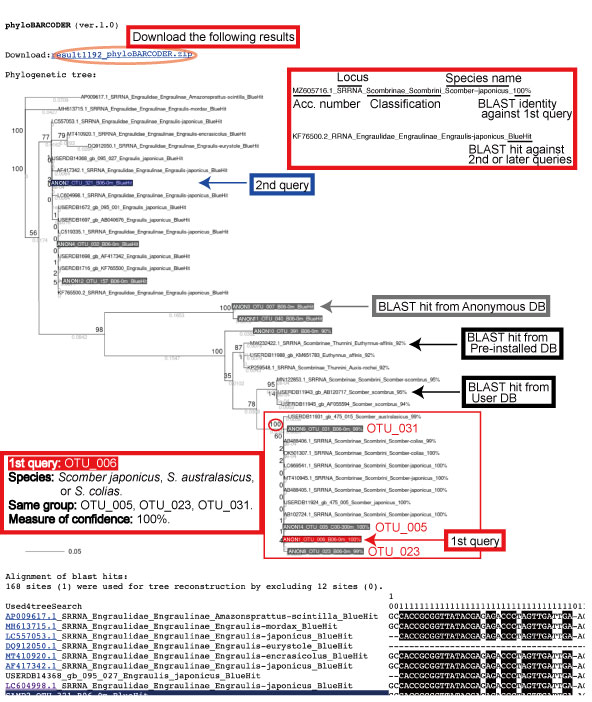
The results can be shown by clicking the link next to Status > Finished.
Above results can be downloaded (result1192_phyloBARCODER.zip). With reference to the estimated phylogenetic tree, the user needs to evaluate species identifications for the queries by sight.
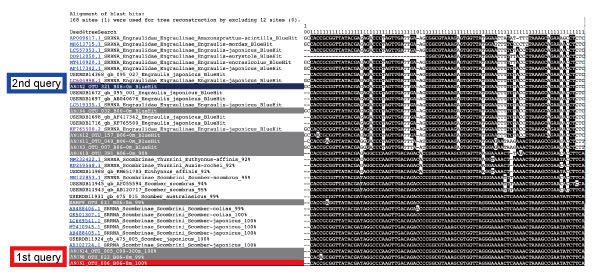
Also, the user can evaluate the species identification from the alignment.
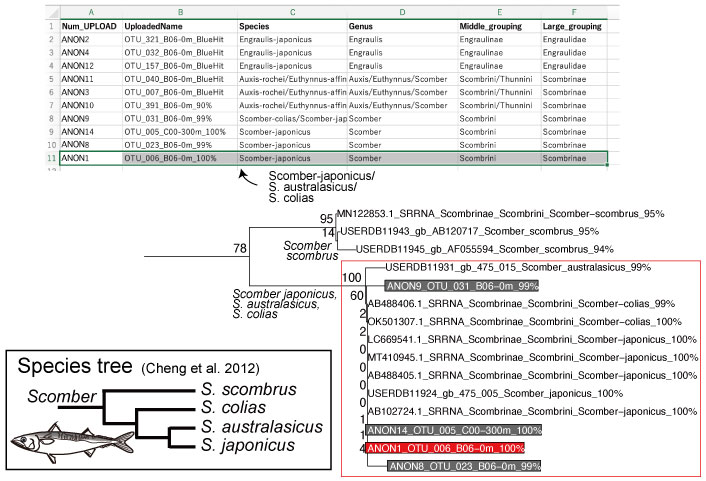
As a species list, phyloBARCODER automatically produces species identifications for the user-defined queries. The species identification and classification* for each query are produced from BLAST hits derived from Pre-installed DB and are saved in the “taxon_assignment_tree.csv” file. For the OTU_6 species name, not only Scomber japonicus but also S. australasicus and S. colias are candidates. To further narrow down the species name, distribution of each species can be considered.
*Those are not produced for BLAST hits from User DB (the custom user reference data).
Example: Fish ASV analysis
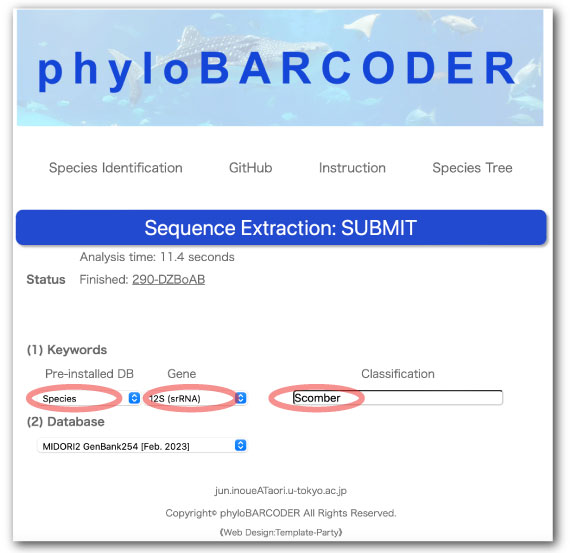
(B) Sequence Extraction
-
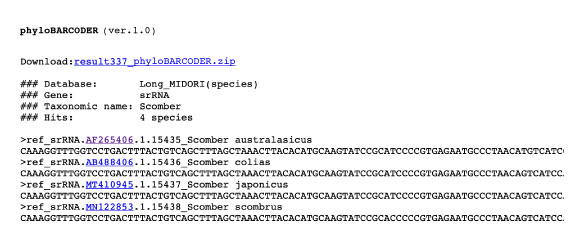
For reference sequences of 12S rRNA gene, we count the number of Scomber species.
Select or enter the following parameters:
- Pre-installed DB
- Species
- Gene
- 12S (srRNA)
- Classification
- Scomber
Citation
-
Inoue J. et al.
phyloBARCODER: A web tool for phylogenetic classification of eukaryote metabarcodes using custom reference databases. Molecular Biology and Evolution 2024, 42: msae111. doi: 10.1093/molbev/msae111
Dependencies
History
| 25/2/5 | v.1.0.6 will be released with BLAST2.10.0 on the server, yurai. | |
| 24/9/18 | v.1.0.5 | MIDORI2 database, GB261, were newly added. |
| 24/7/24 | v.1.0.4 | Reconstructions tree only with all anonymous sequences is revised. See the "Number of queries" option and explanations. |
| 24/6/18 | v.1.0.1 | Published |